GENE SILENCING
RNAi Substrates - shRNA, siRNA & miRNA

RNA Interference (RNAi) is a gene-silencing process that targets mRNA hence lowering protein expression. The key RNAi substrates regulating this process are shRNA, siRNA and miRNA.
TEMA RICERCA provides comprehensive and innovative RNAi tools for various applications:
Key Features:
- Genome wide coverage for human, mouse & rat genes
- Higher potency with minimal interferon response
- Performance guaranteed!
RNA interference (RNAi) tools for gene silencing:
Cloned shRNA
HuSH-29: Genome-wide shRNA for Human, Mouse and Rat Genes
Lentiviral shRNA Now Available!
HuSH-29 is pre-designed shRNA with genome wide coverage of human, mouse and rat. Learn more about HuSH-29 shRNA
TEMA RICERCA also offers custom shRNA service for specific sequence and other species through Exact-shRNA.
Double Gene Knockdown Experiment

Features and Benifits:
-
Lentiviral vector option:
- pGFP-C-shLenti: ideal for primary cells, with GFP reporter
-
Three retroviral vector options:
- pRS: basic retroviral vector without fluorescence
- pRS-GFP: GFP signal to monitor transfection
- pRS-RFP: ideal for double knockdown or co-transfection
- Guaranteed successful gene knockdown (≥70%)
- Versatile applications: transient/stable transfection or retroviral infection
- Delivers 5 μg transfection-ready DNA prepared by PowerPrep® HP Kits
RNAi Duplex Oligos
Overview
RNA interference is a conserved pathway common to plants and mammals where double-stranded RNAs (dsRNAs) suppress expression of genes with complementary sequences [1-2]. RNAi was first observed in lower organisms, such as plants or nematodes. In these systems, long dsRNAs serve as effective triggers of RNAi. Long dsRNAs are not the actual triggers but are degraded by the endoribonuclease Dicer into small effector molecules called siRNAs (small interfering RNAs). SiRNAs are usually around 21 bases long with a central 19 bp duplex and 2-base 3′-overhangs. In mammals, Dicer processing occurs as a complex with the RNA-binding protein TRBP. The nascent siRNA associates with Dicer, TRBP, and Ago2 to form the RNA-Induced Silencing Complex, or RISC, which mediates gene silencing [3]. Once in RISC, one strand of the siRNA (the passenger strand) is degraded or discarded while the other strand (the guide strand) remains to direct sequence specificity of the silencing complex. The Ago2 component of RISC is a ribonuclease that cleaves a target RNA under direction of the guide strand. A schematic overview of the pathways involved in degradative RNAi is shown below.

Although long dsRNAs (several hundred bp) are commonly employed to trigger RNAi in C. elegans or D. melanogaster, these molecules will activate the innate immune system and trigger interferon (IFN) responses in higher organisms. RNAi can be performed in mammalian cells using short RNAs, which generally do not induce IFN responses. Many researchers today employ synthetic 21-mer RNA duplexes as their RNAi reagents, which mimic the natural siRNAs that result from Dicer processing of long substrate RNAs. An alternative approach is to use synthetic RNA duplexes that are greater than 21-mer in length, which are substrates for Dicer.
Antisense Oligonucleotides
Antisense oligonucleotides have been used for over twenty-five years to inhibit gene expression levels both in vitro and in vivo. Recent improvements in design and chemistry of antisense compounds have enabled this technology to become a routinely used tool in basic research, genomics, target validation and drug discovery. It is becoming increasingly popular to confirm phenotypes seen using RNAi by gene silencing antisense DNA oligos. A nucleic acid sequence, usually 18–25 bases long, is designed in antisense orientation to the mRNA of interest; the sequence is made as a synthetic oligonucleotide and is introduced into the cell or organism. Hybridization of the antisense oligo to the target mRNA results in RNase H cleavage of the message which prevents protein translation and thereby blocks gene expression. Antisense oligonucleotides containing a native DNA or phosphorothioate-modified DNA segment of at least six bases long will bind the target mRNA and form an RNA/DNA heteroduplex, which is a substrate for endogenous cellular RNases H [1–2]. The decrease in mRNA levels can be measured using real-time PCR.

Figure 1. Antisense Mediated Cleavage of the Targeted Message by RNase H.
Phosphorothioates and Chimeric Oligos
While unmodified oligodeoxynucleotides can display some antisense activity, they are subject to rapid degradation by nucleases and are therefore of limited utility. The simplest and most widely used nuclease-resistant chemistry available for antisense applications is the phosphorothioate (PS) modification. In phosphorothioates, a sulfur atom replaces a non-bridging oxygen in the oligo phosphate backbone. In the IDT ordering system, an asterisk indicates the presence of a phosphorothioate internucleoside linkage. PS oligos can show greater non-specific protein binding than unmodified phosphodiester (PO) oligos, which can cause toxicity or other artifacts when present at high concentrations. These problems seem to be worst with PS-DNA and PS-DNA/LNA chimeras. Non-specific protein binding of PS oligos can be minimized by making the oligonucleotide as short as possible (thereby reducing PS content) or using chimeric designs with 2’-O-methyl RNA.

LNA, 2′-O-Methyl RNA, and 5-Methyl dC
State-of-the-art antisense design employs chimeras having both DNA and modified-RNA bases. The use of modified RNA, such as 2′-O-methyl (2’OMe) RNA or LNAs (Exiqon) in chimeric antisense designs, increases both nuclease stability and affinity (Tm) of the antisense oligo to the target mRNA [3–5]. These modifications, however, do not activate RNase H cleavage. The preferred antisense design incorporates 2′-O-modified RNA or LNA in chimeric antisense oligos that retain an RNase H activating domain of DNA (or phosphorothioate DNA). As LNA bases confer significant nuclease resistance, we recommend phosphorothioate modification of only the DNA gap, leaving the LNA flanks as phosphodiester linkages in chimeric LNA antisense oligos. For synthesis reasons, a 3′-phosphate is preferred when an LNA base is at the 3′ end.
It can also be beneficial to substitute 5-Methyl-dC for dC in the context of CpG motifs. Substitution of 5-Methyl dC for dC will slightly increase the Tm of the antisense oligo. Use of 5-Methyl dC in CpG motifs can also reduce the chance of adverse immune responses in vivo. IDT recommends that all antisense oligos receive HPLC purification and that oligos undergo a Na+ salt exchange before use in cells or live animals to ensure that salts used in purification are removed.
| Examples of RNase H Active Antisense Oligos | |
|---|---|
| 5′ T*C*C*T*G*C*G*A*A*A*T*G*T*C*C*A*T*C*G*T 3′ | DNA, All PS |
| 5′ T*C*C*TGCGAAATGTCCAT*C*G*T 3′ | DNA, PS/PO chimera |
| 5′ mU*mC*mC*mU*mG*C*G*A*A*A*T*G*T*C*C*mA*mU*mC*mG*mU 3′ | 2′OMe/DNA chimera, All PS |
| 5’ +T+C+C+T+GC*G*A*A*A*T*G*T*C*C+A+T+C+G+T/3Phos/3′ | LNA/DNA chimera, PS/PO chimera, 3′-phos |
* = Phosphorothioate Bonds
mN = 2′-O-Me RNA base
+N = LNA base
References
- Walder RY and Walder JA (1988) Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci USA, 85:50115015.
- Dagle JM, Walder JA, and Weeks DL (1990) Targeted degradation of mRNA in Xenopus oocytes and embryos directed by modified oligonucleotides: studies of An2 and cyclin in embryogenesis. Nucleic Acids Res, 18:47514757.
- Braasch DA, Liu Y, and Corey DR (2002) Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res, 30:51605167.
- Kurreck J, Wyszko E, et al. (2002) Design of antisense oligos stabilized by locked nucleic acids. Nucleic Acids Res, 30:19111918.
- Grunweller A, Wyszko E, et al. (2003) Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2’-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res, 31:31853193.
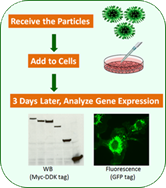
Lenti shRNA transduction particles
Lentiviral system for efficient DNA delivery (Lenti shRNA & LentiORF)
OriGene is licensed to provide lentiviral vectors and related products.
- Third generation Lentiviral system
- Ideal for delivering DNA into hard-to-transfect and animal models
- Replication-incompetent system for safety>
- Improved biosafety features
Both shRNA and ORF lentiviral particles are offered!
- Ready-to-Use: Just add to cells and get results
- Coverage: Genome-wide, Human, Mouse and Rat
- Broad spectrum: Transduce dividing, non-dividing cells and in vivo
- Biosafety: Replication deficient 3rd generation lenti particles