Chromatin & transcription
Chromatin structures are regulated by various mechanisms including histone modification and chromatin remodeling, which involve the binding of transcription factors. By using tools such as chromatin immunoprecipitation, it is possible to gain further insight into the dynamic interactions between transcription proteins and components of chromatin, and to ultimately understand their roles in cellular functions such as gene transcription and epigenetic silencing.
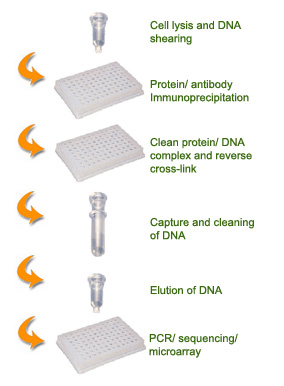
General Chromatin immunoprecipitation (ChIP) is an antibody-based method used for determining the location of DNA binding sites on the genome for a particular protein of interest. This technique is a convenient means for studying protein-DNA interactions that occur inside the nucleus of cells and for understanding cellular processes. Downstream applications of ChIP include ChIP-PCR, ChIP-sequencing, and ChIP-on-chip (microarrays).
By investigating protein-DNA interaction in vitro, it is possible to identify the genetic targets of DNA, which leads to a better understanding of cellular processes. Measurement of direct interactions between protein and DNA in vitro has an advantage in analyzing the binding of different transcription factors to specific DNA consensus sequences located in the gene promoters.
Methyl-Histone Chromatin Immunoprecipitation (ChIP) is a powerful technique for studying protein-DNA interaction in vivo, and can be coupled with various downstream applications to profile and map histone methylation patterns.
Acetyl-Histone Chromatin Immunoprecipitation (ChIP) can be used as a tool to identify activated genes associated with acetylated histones, and can be coupled with qualitative and quantitative PCR, MSP, DNA sequencing, and Southern blot as well as DNA microarrays to further profile or map histone acetylation patterns.
Methyl-DNA Binding Protein Chromatin Immunoprecipitation (ChIP) would be useful in the identification of silenced genes associated with MBD2, and can be combined with qualitative and quantitative PCR, MSP, DNA sequencing, DNA microarrays, and Southern blot to profile or map MBD2 binding patterns. Methyl-CpG-binding domain protein 2 (MBD2) is a member of the MBD protein family and has been shown to catalyze demethylation by direclty removing methyl groups from 5-methylcytosine residues in DNA.
SCHEMATIC PROCEDURE
(e.g. Tri-Methyl-Histone H3K9 ChIP Kit)